
piRNAs - Description
- Small RNAs - An introduction
- piRNAs - A new dimension to gene silencing
- Organization of piRNAs on the genome
Small RNAs - An introduction
Gene expression is mediated by a wide variety of RNA molecules. Messenger RNAs (mRNAs) are protein-coding RNAs, which are involved in translation. There are RNA species produced by eukaryotes, classified as non-coding RNAs, which are not involved in translation but play a major role in regulation of gene expression at the transcriptional, post-transcriptional and translational levels. The small non-coding RNA molecules along with the Argonaute family of proteins have been identified as key players in various forms of sequence specific gene silencing, including RNA interference (RNAi), translational repression and heterochromatin formation. Argonaute proteins are composed of the N-terminal, PAZ (Piwi-Argonaute-Zwille), middle and the C-terminal PIWI (P-element-induced wimpy testis) domains (Tolia et al., 2007).
Five genes have been found to encode distinct members of the Argonaute family of proteins in Drosophila, namely AGO1, AGO2, Aubergine (Aub), Piwi and AGO3 (Gunawardane et al., 2007), of which AGO1 and AGO2 belong to the Argonaute (AGO) subfamily. The remaining three proteins Aub, Piwi and AGO3 occur abundantly in the germline cells and are grouped under the PIWI subfamily of proteins. They play a vital role in the formation of germline cells and also exhibit target RNA cleavage by a process called Slicing, thereby silencing retrotransposons and other repeat elements in the germline.
These proteins act as molecular scaffolds that present the small, guide RNA molecules of RNA-silencing to their complementary targets, by forming a ribonucleoprotein complex. Several types of small RNAs have been classified based on their biogenesis and mode of action. The various classes of small RNAs, their source, length and functions have been listed in Table 1.

Table 1. Different classes of small RNAs
piRNAs - A new dimension to gene silencing
Piwi-interacting RNAs (piRNAs) are a novel class of small RNAs isolated from the mammalian germline cells (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Watanabe et al. 2006). piRNAs interact with the Piwi subfamily of proteins and form a ribonucleoprotein complex called Piwi-interacting RNA complex (piRC), which has been extracted and purified from rat testes (Lau et al., 2006). Rat homologs to Piwi (Riwi) and the human RecQ1 (rRecQ1) proteins were identified from this complex. Due to the presence of the RecQ protein, the piRC exhibits ATP-dependent DNA helicase activity. Figure 1 shows a sketch of piRC, generated by fitting piRNA sequence with the Piwi protein from A. fulgidus (PDB ID 1YTU), using SwissPDB Viewer. Around one million piRNA molecules have been reported per spermatocyte or round spermatid (Aravin et al., 2006). Large scale sequencing of piRNAs from rat, mouse and human testes by different experimental groups have yielded very high number of piRNA sequences. (Girard et al., 2006, Lau et al., 2006, Watanabe et al., 2006, Aravin et al., 2006 and Grivna et al., 2006).
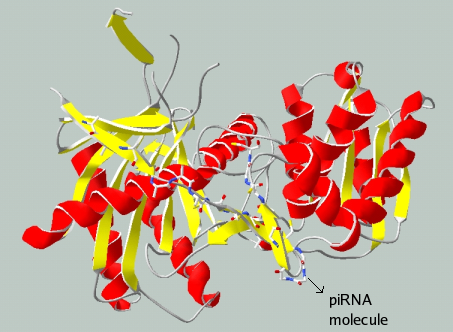
Figure 1. Piwi interacting RNA complex - Generated using SwissPDB Viewer, by fitting piRNA sequence to the crystal structure of PIWI protein (1YTU) from A. fulgidus
Organization of piRNAs on the genome
It has been observed that more than 80% of the entire piRNAs mapped to unique genomic positions. The remaining sequences have been found mapping to multiple genomic loci. Length of piRNA is around 19 – 33 nucleotides, mostly falling in the range of 26–33 nucleotides, while miRNAs and siRNAs are usually ~20-24 bases long. piRNAs identified from the three widely studied organisms namely human, mouse and rat show a strong preference for the 5' uridine residue and they are unevenly distributed among the chromosomes. Further, piRNAs have been found to occur in clusters of length 20 to 100 kilobases, with piRNA density varying between 40 to 4000, most of the clusters being derived from one of the two genomic strands. Such clusters have been named as mono-directional clusters. However, a few bidirectional clusters have also been identified, which are derived from both the strands and is suggestive of possible divergent transcription from the piRNA precursors. Figure 2 shows the schematic of monodirectional and bidirectional clusters in piRNAs.
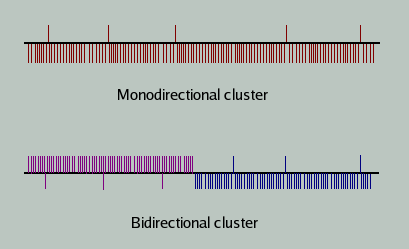
Figure 2. Schematic of monodirectional and bidirectional piRNA clusters